Membrane Proteins
CNS Regeneration
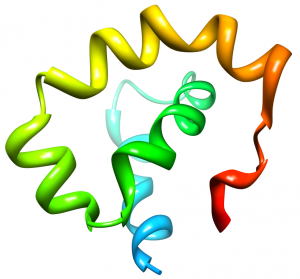 Nogo is a membrane protein known to inhibit axonal growth within the central nervous system (CNS). Disabling Nogo following spinal cord injury or stroke may allow regrowth of damaged axons. One domain of Nogo, termed Nogo-66, is present extracellularly on the oligodentrocyte cells of the CNS. Nogo-66 achieves axonal growth inhibition by binding to the Nogo Receptor (NgR) on neuronal cells. We are interested in characterizing the structure of Nogo-66 and how it interacts with the Nogo Receptor. A detailed understanding of the interaction between Nogo-66 and NgR will prove useful for designing drugs that will interfere with the ligand-receptor interaction and provide recovery from CNS injury.
Nogo is a membrane protein known to inhibit axonal growth within the central nervous system (CNS). Disabling Nogo following spinal cord injury or stroke may allow regrowth of damaged axons. One domain of Nogo, termed Nogo-66, is present extracellularly on the oligodentrocyte cells of the CNS. Nogo-66 achieves axonal growth inhibition by binding to the Nogo Receptor (NgR) on neuronal cells. We are interested in characterizing the structure of Nogo-66 and how it interacts with the Nogo Receptor. A detailed understanding of the interaction between Nogo-66 and NgR will prove useful for designing drugs that will interfere with the ligand-receptor interaction and provide recovery from CNS injury.
Our Nogo structure paper: Vasudevan, SV, Schulz, J, Zhou, C and Cocco, MJ*. (2010) Protein Folding at the Membrane Interface, the Structure of Nogo-66. Proceedings of the National Academy of Sciences, 107:6847-51
Vaccine development
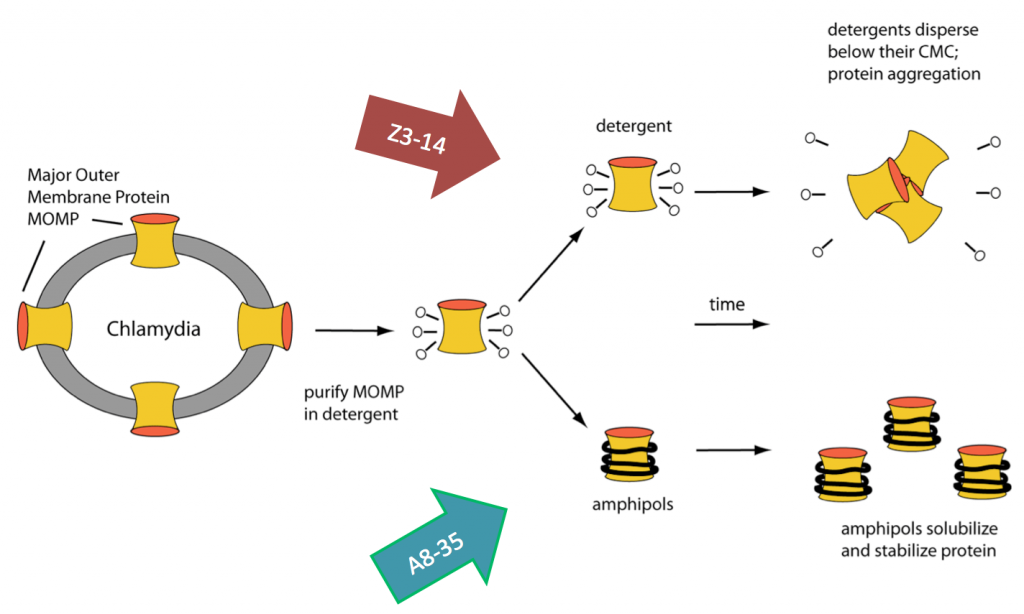
Chlamydia trachomatis is one of the most common bacterial pathogens found in all regions of the World. Chlamydia vaccines formulated with whole inactivated and viable organisms were tested in the 1960’s in humans and in non-human primates to protect against trachoma. Some vaccine protocols induced protection, but the latter lasted only 1–2 years. Furthermore, after re-exposure to Chlamydia, some of the immunized individuals developed a hypersensitivity reaction, most likely attributed to an antigenic component of the bacteria. Consequently, the need to develop a subunit vaccine was considered. A good candidate for a subunit vaccine is the C. trachomatis major outer membrane protein, MOMP. Integral membrane proteins are kept soluble in aqueous solutions using detergents and MOMP/detergent vaccine formulations have been used with some success. We have improved on these vaccines by solubilizing MOMP in amphiboles (a novel class of amphipathic polymers). In collaboration with Professor Luis de la Maza (UCI pathology) and Professor Jean-Luc Popot (CNRS, Paris), we have shown for the first time that a vaccine formulated with APols and nMOMP, an integral membrane protein, induces a more robust protective immune response than the same antigen formulated with a detergent. Moreover, the vaccine formulation is stable at room temperature for months.
Our manuscript describing the development of vaccine formulations using a membrane protein in amphiboles: Tifrea, DF, Sun, G, Pal, S, Zardeneta1, G, Cocco, MJ, Popot, J-L, de la Maza, LM (2011) Amphipols stabilize the Chlamydia major outer membrane protein and enhance vaccine protection based on a membrane-protein formulation. Vaccine 29:4623-31
Natural Antibiotics
 Defensins are family of antimicrobial peptides having activity against a range of microorganisms: gram-positive and gram-negative bacteria, fungi and some viruses. In addition to their antimicrobial activity, emerging evidence suggests that they can also assume fundamental roles in both innate and adaptive immunity.
Defensins are family of antimicrobial peptides having activity against a range of microorganisms: gram-positive and gram-negative bacteria, fungi and some viruses. In addition to their antimicrobial activity, emerging evidence suggests that they can also assume fundamental roles in both innate and adaptive immunity.
More than 300 defensins have been identified to date and they are represented in a range of organisms including mammals, birds, invertebrates, plants and recently in the ebony-cup fungus. Our lab focuses on structural characterization of some of these defensins and their interactions with membranes to understand their mode of action.
Our defensin structure paper: Vasudevan, S, Yuan, J, Ösapay, G, Tran, P, Tai, K, Liang, W, Kumar, V, Selsted, ME*, and Cocco, MJ* (2008) Synthesis, Structure and Activities of an Oral Mucosal Alpha-Defensin from Rhesus Macaque, Journal of Biological Chemistry, 283:35869-77
DNA Binding Proteins
Gene Regulation
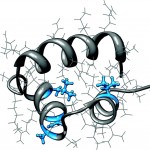 CytR is a bacterial transcription factor that represses production of the genes of the cytidine regulation pathway. While containing high sequence and functional homology to LacR, the lactose repressor, CytR has unique DNA sequence recognition ability in its dimeric form. While many DNA binding proteins form dimers to bind to palindromic sequences with a defined spacing, only CytR has been shown to have the ability to bind to half sites separated by a varying number of bases. Our NMR studies allow us to elucidate the structure and dynamics of the protein as it binds DNA. In collaboration with the Senear lab, we are working toward a greater understanding of the mechanism of gene recognition and expression regulation in general.
CytR is a bacterial transcription factor that represses production of the genes of the cytidine regulation pathway. While containing high sequence and functional homology to LacR, the lactose repressor, CytR has unique DNA sequence recognition ability in its dimeric form. While many DNA binding proteins form dimers to bind to palindromic sequences with a defined spacing, only CytR has been shown to have the ability to bind to half sites separated by a varying number of bases. Our NMR studies allow us to elucidate the structure and dynamics of the protein as it binds DNA. In collaboration with the Senear lab, we are working toward a greater understanding of the mechanism of gene recognition and expression regulation in general.
Our paper describing the structure of the CytR DBD: Moody, CL, Tretyachenko-Ladokhina, V, Laue, T., Senear, DF and Cocco, MJ* (2011) Multiple Conformations of the Cytidine Repressor DNA-Binding Domain Coalesce to One Upon Recognition of a Specific DNA Surface. Biochemistry, 50:6622-32